Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Sogabe, Tomochika*; Nakagawa, Hiroshi; Yamada, Takeshi*; Koseki, Shigenobu*; Kawai, Kiyoshi*
Biophysical Journal, 121(20), p.3874 - 3882, 2022/10
Times Cited Count:2 Percentile:19.58(Biophysics)The purpose of this study was to clarify the glass transition behavior of bacteria (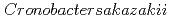 ) as a function of water activity (
) as a function of water activity (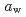 ). Mechanical relaxation was investigated at 298 K, and the mechanical
). Mechanical relaxation was investigated at 298 K, and the mechanical 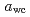 (
(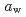 at which mechanical glass transition occurs at 298 K) was determined to be 0.667. Temperature-dependency of mean square displacement was investigated by inelastic neutron scattering. From the linear fitting, two dynamical transition temperatures (low and high-
at which mechanical glass transition occurs at 298 K) was determined to be 0.667. Temperature-dependency of mean square displacement was investigated by inelastic neutron scattering. From the linear fitting, two dynamical transition temperatures (low and high-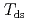 ) were determined. There was a minor effect of
) were determined. There was a minor effect of 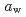 on the low-
on the low-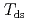 except for the anhydrous sample. The high-
except for the anhydrous sample. The high-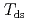 largely increased with the decrease in
largely increased with the decrease in 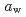 . The dynamical
. The dynamical 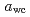 determined by high-
determined by high-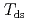 (0.688) was slightly higher than the mechanical
(0.688) was slightly higher than the mechanical 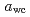 because of the difference in the measurement time-scale. The high-
because of the difference in the measurement time-scale. The high-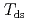 was converted to the glass transition temperature (
was converted to the glass transition temperature (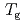 ), and anhydrous
), and anhydrous 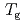 was estimated to be 411 K. Bacterial inactive-active transition was discussed according to the glass transition behavior.
was estimated to be 411 K. Bacterial inactive-active transition was discussed according to the glass transition behavior.
 -casein micelles by neutron spectroscopy
-casein micelles by neutron spectroscopyNakagawa, Hiroshi; Appavou, M.-S.*; Wuttke, J.*; Zamponi, M.*; Holderer, O.*; Schrader, T. E.*; Richter, D.*; Doster, W.*
Biophysical Journal, 120(23), p.5408 - 5420, 2021/12
Times Cited Count:1 Percentile:8.05(Biophysics)Casein proteins are characterized by its extended structural features and its dynamics, which are different from those of typical folded proteins, due to a large number of proline residues. We were able to describe the unique structural dynamics by Orenstein-Uhlenbeck Brownian oscillator model by several types of QENS experiments with wide time-scales. The characterized dynamics also allowed us to discuss the effective sequestration of calcium phosphate in terms of its unique dynamical structure. The biological role of the physical processes of the proteins on the nanosecond order on the biological functions is presented.
Nakagawa, Hiroshi; Saio, Tomohide*; Nagao, Michihiro*; Inoue, Rintaro*; Sugiyama, Masaaki*; Ajito, Satoshi; Tominaga, Taiki*; Kawakita, Yukinobu
Biophysical Journal, 120(16), p.3341 - 3354, 2021/08
Times Cited Count:4 Percentile:38.66(Biophysics)A multi-domain protein can have various conformations in solution. Interactions with other molecules result in the stabilization of one of the conformations and change in the domain dynamics. SAXS, a well-established experimental technique, can be employed to elucidate the conformation of a multi-domain protein in solution. NSE spectroscopy is a promising technique for recording the domain dynamics in nanosecond and nanometer scale. Despite the great efforts, there are still under development. Thus, we quantitatively removed the contribution of diffusion dynamics and hydrodynamic interactions from the NSE data via incoherent scattering, revealing the differences in the domain dynamics of the three functional states of a multi-domain protein, MurD. The differences among the three states can be explained by two domain modes.
 electron paramagnetic resonance
electron paramagnetic resonanceSaio, Tomohide*; Hiramatsu, Soya*; Asada, Mizue*; Nakagawa, Hiroshi; Shimizu, Kazumi*; Kumeta, Hiroyuki*; Nakamura, Toshikazu*; Ishimori, Koichiro*
Biophysical Journal, 120(15), p.2943 - 2951, 2021/08
Times Cited Count:1 Percentile:8.05(Biophysics)A rigid double-arm lanthanide tag was utilized in electron paramagnetic resonance spectroscopy to measure the distance between two specific points on a protein, and conformational states and distribution of a multi-domain protein enzyme MurD was investigated. Although the previous crystallographic and NMR studies have reported the three distinct conformational states of MurD, our data unveiled that the protein exists in much more variety of conformational states in the absence of the ligand. Given the fact that MurD is one of the potent drug target for infectious diseases, the finding in this study will provide important structural basis for drug development.
Nakagawa, Hiroshi; Jochi, Yasumasa*; Kitao, Akio*; Yamamuro, Osamu*; Kataoka, Mikio*
Biophysical Journal, 117(2), p.229 - 238, 2019/07
Times Cited Count:3 Percentile:13.03(Biophysics)Softness and rigidity of proteins are reflected in the structural dynamics, which are in turn affected by the environment. The characteristic low-frequency vibrational spectrum of a protein, known as boson peak, is an indication of the structural rigidity of the protein at cryogenic temperature or dehydrated conditions. In this paper, the effect of hydration, temperature, and pressure on the boson peak and volumetric properties of a globular protein are evaluated by using inelastic neutron scattering and molecular dynamics simulation. Hydration, pressurization, and cooling shift the boson peak position to higher energy and depress the peak intensity and decreases the protein and cavity volumes, although pressure hardly affects the boson peak of the fully hydrated protein. A decrease of each volume means the increase of rigidity, which is the origin of the boson peak shift. The boson peak profile can be predicted by the total cavity volume. This prediction is effective for the evaluation of the net quasielastic scattering of incoherent neutron scattering spectra when the boson peak cannot be distinguished experimentally because of a strong contribution from quasielastic scattering.
Yang, L.-W.*; Kitao, Akio*; Huang, B.-C.*; Go, Nobuhiro*
Biophysical Journal, 107(6), p.1415 - 1425, 2014/09
Times Cited Count:18 Percentile:54.3(Biophysics)Oikawa, Masataka; Yonetani, Yoshiteru
Biophysical Journal, 98(12), p.2974 - 2983, 2010/06
Times Cited Count:9 Percentile:23.53(Biophysics)Yonetani, Yoshiteru; Kono, Hidetoshi
Biophysical Journal, 97(4), p.1138 - 1147, 2009/08
Times Cited Count:28 Percentile:61.07(Biophysics)Ishida, Hisashi; Hayward, S.*
Biophysical Journal, 95(12), p.5962 - 5973, 2008/12
Times Cited Count:29 Percentile:60.62(Biophysics)Matsumoto, Atsushi; Kamata, Tetsuji*; Takagi, Junichi*; Iwasaki, Kenji*; Yura, Kei
Biophysical Journal, 95(6), p.2895 - 2908, 2008/09
Times Cited Count:19 Percentile:44.72(Biophysics)Protein is activated, but the activation mechanism and generality of the conformational change remain to be elucidated. We performed normal mode analysis of the elastic network model on integrin 
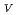

 ectodomain in the bent form and identified key residues which were influential on molecular motions. Iterative normal mode calculations demonstrated that the specific non-bonded interactions involving the key residues work as a snap to keep integrin in the bent form. The importance of the key residues for the conformational change was further verified by mutation experiments. Conservation pattern of amino acid residues among integrin family showed that the characteristic pattern of residues seen around these key residues is found in the limited groups of integrin
ectodomain in the bent form and identified key residues which were influential on molecular motions. Iterative normal mode calculations demonstrated that the specific non-bonded interactions involving the key residues work as a snap to keep integrin in the bent form. The importance of the key residues for the conformational change was further verified by mutation experiments. Conservation pattern of amino acid residues among integrin family showed that the characteristic pattern of residues seen around these key residues is found in the limited groups of integrin  chains.
chains.
Nakagawa, Hiroshi; Jochi, Yasumasa*; Kitao, Akio*; Kataoka, Mikio
Biophysical Journal, 95(6), p.2916 - 2923, 2008/09
Times Cited Count:49 Percentile:78.26(Biophysics)Jochi, Yasumasa*; Nakagawa, Hiroshi; Kataoka, Mikio; Kitao, Akio*
Biophysical Journal, 94(11), p.4435 - 4443, 2008/06
Times Cited Count:24 Percentile:53.62(Biophysics)Hydration effects on protein dynamics were investigated by comparing the frequency dependence of the calculated neutron scattering spectra between full and minimal hydration states at temperatures between 100 and 300 K. The protein boson peak is observed in the frequency range 1-4 meV at 100 K in both states. The peak frequency in the minimal hydration state shifts to lower than that in the full hydration state. Protein motions with frequency higher than 4 meV were shown to undergo almost harmonic motion in both states at all temperatures simulated, whereas those with frequency lower than 1 meV dominate the total fluctuations above 220 K and contribute to the origin of the glass-like transition. At 300 K, the boson peak becomes buried in the quasi-elastic contributions in the full hydration state, but is still observed in the minimal hydration state. The boson peak is observed when protein dynamics are trapped within a local minimum of its energy surface. Protein motions, which contribute to the boson peak, are distributed throughout the whole protein. Fine structure of the dynamics structure factor is expected to be detected by the experiment if a high resolution instrument is developed in the near future.
Fujiwara, Satoru; Plazanet, M.*; Matsumoto, Fumiko; Oda, Toshiro*
Biophysical Journal, 94(12), p.4880 - 4889, 2008/06
Times Cited Count:9 Percentile:21.55(Biophysics)Actin plays crucial roles in various aspects of cell motility. Flexibility of F-actin, a filamentous polymer formed by polymerization of the monomers (G-actin), is important for such a variety of functions. This flexibility allows F-actin to interact with various proteins, thereby expressing multiple functions. Understanding the variety of functions of actin thus requires understanding the flexibility of F-actin. As a first step towards this ultimate purpose, we carried out elastic incoherent neutron scattering (EINS) experiments on G-actin and F-actin under hydrated states. The mean square displacement (MSD) was estimated from the EINS measurements. Temperature dependence of MSD showed that two dynamical transitions occur at about 150 K and about 245 K, and that behavior of MSD is different between G-actin and F-actin, such that G-actin is "softer" than F-actin. The different behavior observed is ascribed to the differences in dynamical heterogeneity between F-actin and G-actin.
Miyazawa, Yuji*; Nishioka, Hirotaka*; Yura, Kei; Yamato, Takahisa*
Biophysical Journal, 94(6), p.2194 - 2203, 2008/03
Times Cited Count:22 Percentile:50.14(Biophysics)DNA photolyase recognizes ultraviolet-damaged DNA and breaks improperly formed covalent bonds within the cyclobutane pyrimidine dimer. Theoretical analysis of the electron-tunneling pathways of the DNA photolyase derived from Anacystis nidulans can reveal the active role of the protein environment in the electron transfer reaction. Here, we report the unexpectedly important role of the single methionine residue, Met-353, where busy trafficking of electron-tunneling currents is observed. The amino acid conservation pattern of Met-353 in the homologous sequences perfectly correlates with experimentally verified annotation as photolyases. The bioinformatics sequence analysis also suggests that the residue plays a pivotal role in biological function. Consistent findings from different disciplines of computational biology strongly suggest the pivotal role of Met-353 in the biological function of DNA photolyase.
Sakamoto, Fuminori; Suzuki, Eiji*; Fujii, Yuki*
Journal of Biochemical and Biophysical Methods, 52(2), p.97 - 109, 2002/07
Times Cited Count:4 Percentile:9.68(Biochemical Research Methods)no abstracts in English